Molecular Basis of Inheritance | CBSE Biology Class XII Notes
THE DNA (Deoxyribonucleic Acid):
- DNA is a long polymer of deoxyribonucleotides.
- The length of the DNA depends on the number of nucleotide base pair present in it.
- Bacteriophage ø×174 has 5386 nucleotides.
- Bacteriophage lambda has 48502 base pairs.
- Escherichia coli have 4.6 X 106 base pairs.
- Human genome (haploid) consists of 3.3 × 109 base pairs.
Structure of Polynucleotide Chain
- DNA and RNA are referred to as Polynucleotide chain
- Nucleotide: Nucleoside + Phosphate moiety (linked via Phosphodiester bond)
- Nucleoside: Pentose Sugar + Nitrogenous base (linked via the N-Glycisidic Bond)
- Pentose sugar is Ribose in RNA and Deoxyribose in DNA.
- Nitrogenous base are two types
- Purine (Adenine and Guanine)
- Pyrimidine (Cytosine, Thymine and Uracil)
- Uracil is found only in RNA in place of Thymine.
- Number of nucleotides joins together through 3’ – 5’ phosphodiester bond to form the polynucleotide chain.
- Polynucleotide chain has a free phosphate moiety at 5’ end of sugar, is referred to as 5’ end and a 3’-OH group on the other end called 3’ end.
- Sugar and phosphate forms the backbone of polynucleotide chain.
Discovery of DNA and determination of its structure
- Friedrich Meischer: Discovered the DNA in 1869 and named it as ‘Nuclein’.
- Wilkins & Franklin : Produced the X-ray diffraction data for DNA structure.
- Watson & Crick : Proposed double helix structure model for DNA based on X-ray diffraction data.
Chargaff’s Rule
- For a double stranded DNA (dsDNA), the ratio between Adenine and Thymine, and Guanine and Cytosine are constant and are equal to one.
Salient features of the Double-helix structure of DNA
- Two polynucleotide chains are coiled to form a double helix. Sugar-phosphate forms backbone for the helix. The bases project in wards to each other.
- The two chains have antiparallel polarity.
- The nitrogen bases of the two strands are paired by Hydrogen bonding. A purine always pairs with a pyrimidine. A = T and C Ξ G.
- The plane of one base pair stacks over the other. This provides stability to the helix along with hydrogen bonding.
- The helix is right handed.
- Pitch (distance parallel to helix that corresponds to one turn of 3600) : 3.4 nm or 34Å.
- Number of bases per turn: 10
- Distance between two adjacent base pair: 0.34 nm.
Central Dogma of Molecular Biology
- Proposed by Francis Crick.
- Talks about the direction of flow of genetic information.
- DNA→ RNA→ Protein
Packaging of DNA Helix
- There is 6.6×109 bp per cell in mammals. Taking 0.34 nm as the distance between consecutive bp, the total length of DNA happens to be 2.2 meters. (6.6 X109 bp X 0.34 X10-9m=2.2 meters)
- 2.2 m of DNA is too large to be accommodated in the nucleus with a dimension of 10−6m.
Packaging of DNA in Prokaryotes (E.g. E. coli)
- Prokaryotes lack a well-defined nucleus.
- Genetic material is scattered in the cytoplasm.
- Nucleoid: The region where DNA being negatively charged (due to phosphate moiety) is associated with protein that is positively charged.
Packaging of DNA in Eukaryotes
- Histones: Positively charged, basic protein.
- Histones are rich in basic amino acids like Lysine and arginine that give it positive charge.
- Histone octamer: Unit of eight molecules of histone.
- DNA (negatively charged) wraps around histone (positively charged) to form nucleosomes.
- 1 nucleosome has approximately 200 bp of DNA.
- Nucleosomes in a chromatin resemble beads present on strings.
- Beads on string structure in chromatin are further packaged to form chromatin fibres, which further coil and condense to form chromosomes during metaphase.
- Non-histone chromosomal (NHC) proteins − Additional set of proteins required for packaging of chromatin at higher level
Chromatin Types
- Euchromatin:
- Lightly Stained.
- Loosely coiled region of chromatin.
- Transcriptionally active.
- Heterochromatin:
- Darkly Stained.
- Tightly coiled region of chromatin.
- Transcriptionally inactive.
The search for genetic material
Transforming Principle
- Experiments performed by Griffith on Streptococcus pneumonia.
- S. pneumonia has two strains: R strain and S strain.
| S Strain | R Strain |
| Produces a smooth colony on culture plate. | Produces a rough colony on culture plate. |
| Produces a polysaccharide coat. | Polysaccharide coat absent. |
| Virulent | Non-Virulent |
- Experiment performed by Griffith:
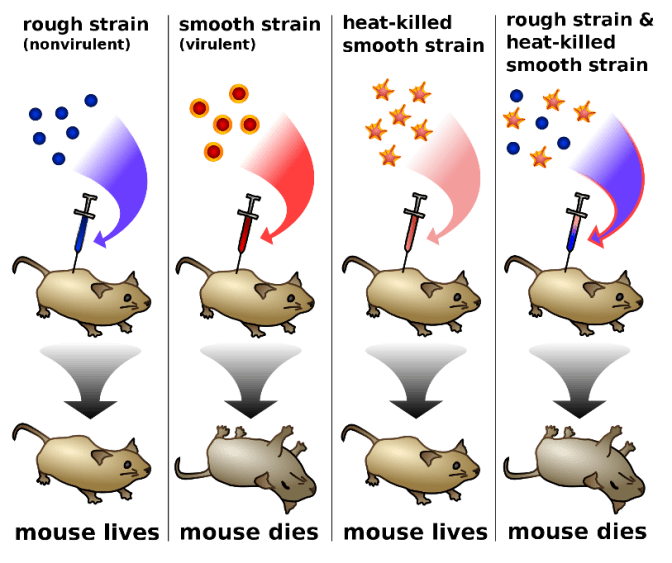
- Live R strain in the presence of heat-killed S strain should not have killed the mouse.
- Somehow the bacteria produce virulence.
- This is because somehow the R strain bacteria are transformed by heat-killed S strain bacteria.
- The transformation must be due to transfer of genetic material.
Biochemical Nature of Transforming Material
- Avery, MacLeod and McCarty worked to determine the biochemical nature of ‘transforming principle’ in Griffith’s experiment.
- They tried transforming the R cells to S cells by using biochemical (proteins, DNA, RNA, etc.) extracted from the S cell.
- They concluded that DNA is the genetic material, as only it could transform the bacterial strain.
DNA as the Genetic Material
- The proof came from the experiment performed by Hersey and Chase.
- They used bacteriophage-Virus that infects bacteria.
- Upon infection the bacteriophage injects its DNA to the host cell and gets integrated to the host genome and subsequently produces more viral particles using the host machineries.
- They experimented to find out whether protein or the DNA that entered the bacterial cell.
Hersey and Chase Experiment
- They grew some viruses on a medium that contained radioactive phosphorus (32P) and some others on medium that contained radioactive sulphur (35S).
- Media containing radioactive Phosphorous had radioactive DNA.
- Media containing radioactive sulphur had radioactive protein.
- Infection: These radioactive bacteriophages were used to infect E. coli. Phage transfers the genetic material to the bacterial cell.
- Blending: Viral coat were separated from the bacterial cell by agitating them in a blender.
- Centrifugation: Viral particles were separated from the bacterial cell by spinning them in a centrifuge machine.
Observation:
- Bacteria infected with phage with radioactive protein (35S)
- No radioactivity detected in cell
- Radioactivity detected in the supernatant.
- Bacteria infected with phage with radioactive DNA (32P)
- Radioactivity detected in cell
- No-radioactivity detected in the supernatant.
Conclusion:
- The above observation concluded that it is the DNA that entered the bacteria from phage and not proteins.
- Hence, it was concluded that DNA is the genetic material and not the protein.
Properties of Genetic Material (DNA and RNA)
- Criteria for a biomolecule to be genetic material:
- It should be able to make copy of it self (Replicate).
- It should be structurally and chemically stable.
- It should all the scope for mutation (essential for the process of evolution).
- It should express itself following the Mendelian principles of inheritance.
Stability of RNA
- The 2′ OH group in RNA is present at every nucleotide makes RNA unstable and degradable.
- RNA also acts as catalyst (ribozyme), hence reactive.
- RNA mutates faster compared to DNA, as it is unstable.
Stability of DNA
- The complementarity of the two DNA strands provides stability to the molecule.
- Thymine instead of uracil in DNA provides additional stability.
- DNA being more stable chemically and structurally is the preferred nucleic acid for storage of genetic material.
RNA World
- RNA was the first genetic material and essential life processes evolved around it.
- DNA has evolved from RNA with chemical modification that makes it more stable.
DNA Replication
- During replication two strands of DNA separate and act as template for the synthesis of new DNA strand that are complementary.
- Semiconservative DNA replication: After one complete replication cycle, each DNA molecule consists of a parental DNA strand and a newly synthesised strand.
Proof of Semiconservative nature (Meselson and Stahl’s Experiment):
- Experiment performed by Meselson and Stahl, on E. coli.
- They used heavier isotope of nitrogen 15N in the media as the only nitrogen source for many generation
- 15N gets incorporated into the newly synthesized DNA (heavy DNA).
- The E. coli then were allowed to grow on a medium with 14N as the nitrogen source.
- Samples were taken at definite time intervals (20 minutes/one generation) as the cells multiplied and the DNA was extracted.
- Heavy DNA can be separated from the normal DNA by CsCl density gradient.
- Various samples were centrifuged independently using Cscl Density gradient centrifugation.
Observation:
- The DNA that was extracted from the culture before transferring to the 15N medium had a heavy density.
- DNA extracted from the culture one generation after the transfer from 15N to 14N had a hybrid or intermediate density.
- DNA extracted after another generation was composed of equal amounts of this hybrid DNA and of ‘light’ DNA.
Conclusion:
- Form the above observations they concluded that DNA replication is semi-conservative in nature.
Mechanism of DNA replication
- The DNA replication occurs in the S phase of cell cycle.
- The main enzyme involved is the DNA dependent DNA polymerase.
- Deoxyribonucleoside triphosphates (dNTPs) serve dual purposes:
- Acts as the substrate
- Provides energy the polymerisation reaction
- DNA replication begins at specific site called the origin of replication.
- Replication fork: During replication the two DNA strand do not completely open up. Instead a small portion is opened up where replication occurs. This site is called replication fork.
- DNA polymerase polymerises the new strand only in one direction, i.e. 5’→3’.
- Continuous/ leading strand: One of the DNA strand with polarity 3’→5’ that acts as template for the new strand synthesis, synthesises the strand continuously.
- Discontinuous/ lagging strand: The other strand with polarity 5’→3’, acting as the template synthesizes the new strand in fragments (Okazaki fragments) that are later joined together by the enzyme DNA ligase.
Transcription
- The process of formation of RNA from DNA is referred to as transcription.
- Only a segment of DNA from only one of the two strands participates in the process of transcription.
- Both strands are not copied during transcription because of the following:
- If both strands get transcribed at the same time then two RNA molecules with different sequences will be formed, and in turn if they code for protein, two different sequences of amino acid would be formed, which in turn give rise to two different proteins. Therefore, one DNA fragment would end up giving rise to two different proteins.
- Two RNA molecules so formed will be complementary to each other, hence would end up forming a double-stranded RNA leaving the entire process of transcription futile.
Transcriptional Unit
- A transcriptional unit has primarily three regions:
- Promoter − Marks the beginning of transcription; RNA polymerase binding site
- Structural gene − Part of the DNA that is actually transcribed
- Terminator − Marks the end of transcription
Structural gene: Template/coding strand
- Template strand: DNA strand with polarity 3’→5’
- Coding strand: DNA strand with polarity 5’→3’
- The sequence of RNA is same with that of the Coding strand.
- All the reference point while defining a transcription unit is made with coding strand.
Promoter:
- The region of DNA where RNA polymerase binds.
- Located towards the 5’ end or upstream region of structural gene.
Terminator:
- Region of DNA that defines the end of transcription.
- Located towards the 3’ end or downstream region of the structural gene.
- This is the site where the termination factor (ρ factor) binds to the RNA polymerase.
Transcription Unit and the Gene
- Gene: The DNA sequence which codes for tRNA or rRNA molecule.
- Cistron: Segment of DNA that contains the genetic code for a single polypeptide.
- The structural genes could be of two types:
- Monocistronic (mostly in eukaryotes)
- Polycistronic (mostly in prokaryotes)
- Monocistronic genes have two parts:
- Exon: Sequences that code for a particular character and is expressed in a matured and processed mRNA.
- Intron: Interrupting sequences that do not appear in a mature and processed mRNA.
Types of RNA
- mRNA (messenger RNA) : It serves as a template for protein synthesis.
- tRNA (transfer RNA) : It brings amino acids during translation and reads the genetic code. Possess the anti-codon.
- rRNA (ribosomal RNA) : They play a structural and catalytic role during translation.
Process of Transcription:
- Transcription has three steps − initiation, elongation, and termination.
Initiation
- RNA polymerase binds to promoter and start transcription.
- The process is catalysed by the DNA dependent RNA polymerase.
- It associates transiently with initiation-factor (σ) to initiate the process.
Elongation
- RNA polymerase uses nucleoside triphosphates (NTPs) as substrate to polymerise the new strand following the rule of complementarity.
Termination
- On reaching the terminator region, the RNA polymerase associates with the termination factor (ρ).
- As a result of this the nascent RNA falls off, so also the RNA polymerase.
- This results into the termination of the process.
Prokaryotic Transcription
- mRNA does not require any processing to become active.
- As there is no separate nucleus in prokaryotes, translation can begin before transcription is completed.
- Thus, in prokaryotes, transcription and translation are coupled.
Eukaryotic Transcription
- Three RNA polymerases are present
- RNA polymerase I transcribes rRNA (28S, 18S and 5.8S)
- RNA polymerase II transcribes hnRNA (mRNA precursor).
- RNA polymerase III transcribes tRNA, snRNA, and 5s rRNA.
- Processing of hnRNA:
- Heterogeneous nuclear RNA (hnRNA) containts both introns and exons.
- RNA splicing: Introns are removed by this process and Exons are joined together.
- Capping: A methyl guanosine triphosphate residue is added to the 5′ end of hnRNA.
- Tailing: 200-300 adenylate residues are added to the 3′ end of hnRNA.
- Fully processed hnRNA, now called mRNA, that is transported out of the nucleus for translation.
Genetic Code
- Genetic code that directs the sequence of amino acids during synthesis of proteins.
- Change in nucleic acid is responsible for change in amino acids in proteins.
- George Gamow: Proposed that in order to code for all the 20 amino acids, the code should be made up of 3 nucleotides.
- Codon: Sequence of three nucleotides that corresponds to a specific amino acid or stop signal during protein synthesis.
- Har Gobind Khorana: gave rise to the checker-board for genetic code.
- The salient features of genetic code are as follows:
- The codon is triplet. 61 codons code for amino acids and 3 codons do not code for any amino acids (stop codons).
- Unambiguous: One codon codes for only one amino acid.
- Degenerate: Some amino acids are coded by more than one codon.
- No punctuations: The codon is read in mRNA in a contiguous fashion.
- Universal: One codon codes for the same amino acid in almost all the species.
- AUG has dual functions. It codes for Methionine and also acts as initiator codon.
Mutations and Genetic Code
- Mutations include insertions, deletions, and rearrangements.
- Mutation results in changed phenotype and diseases such as sickle cell anaemia. (Change Glu→ Val in gene coding for β-globin chain of haemoglobin).
- Insertion or deletion of a single base pair disturbs the entire reading frame in mRNA. Such mutations are called frameshift mutations.
- Frameshift mutations hold the proof of the fact that codon is triplet because if we insert three or multiple of three bases followed by the deletion of same number of bases, then the reading frame will remain unaltered.
tRNA-The Adapter Molecule
- tRNA is the adapter molecule that on one hand read the code and on other hand bind to specific amino acid.
- Anticodon loop: It has bases complementary to the code
- Amino acid acceptor end: Contains amino acid binding site.
- tRNAs are specific for each amino acid.
- Initiator tRNA: It carries formylated metheionine (f-met). It helps in initiation of translation.
- No tRNA for stop codon.
- The secondary structure of tRNA looks like a clover-leaf.
Translation
- It is the process where polypeptide chains (proteins) are formed from a mRNA.
- Amino acids are polymerised (joined by peptide bond) to form a polypeptide.
- At first charging of tRNA (amino-acylation of tRNA) takes place. In this, amino acids are activated in the presence of ATP and are linked to their corresponding tRNA. This energetically favours the formation of peptide bond between two amino acids.
- Translation occurs in Ribosome. Ribosomes have 2 subunits: a large subunit and a small subunit.
- Smaller subunit comes in contact with mRNA to initiate the process of translation.
- Translational unit in an mRNA is the region flanked by start codon and stop codon.
- Untranslated regions (UTR) are the regions on mRNA that are not translated, but are required for efficient translation process. They are present before start codon (5′ UTR) or after stop codon (3′ UTR).
- Initiation: Initiator tRNA recognises the start codon.
- Elongation: The t-RNA-amino acid complexes bind to their corresponding codon on the mRNA and base pairing occurs between codon on mRNA and tRNA anticodon. tRNA moves from codon to codon on the mRNA and amino acids are added one by one.
- Termination: Release factor binds to stop codon to terminate the translation.
Regulation of gene expression
- In eukaryotes, the regulation could be exerted at:
- transcriptional level (formation of primary transcript),
- processing level (regulation of splicing),
- transport of mRNA from nucleus to the cytoplasm,
- translational level
- The metabolic, physiological or environmental conditions that regulate the expression of genes.
Prokaryotic regulation of gene expression
- Gene expression is regulated by controlling the rate of transcriptional initiation.
- The activity of RNA polymerase at a given promoter is regulated by accessory proteins. The accessory proteins affect the ability of a promoter to recognise start sites.
- A regulatory protein could be activator or repressor.
- Accessibility of promoter is also affected by operators. Operator is the region located adjacent to promoter.
- Operon– a polycistronic structural gene is regulated by a common promoter and regulatory genes.
- Each operon has a specific operator and a specific repressor.
- Usually operator binds to a repressor protein.
Lac Operon
- Lac operon was first described by Jacob and Monad.
- Bacteria generally prefer glucose as the carbon source to obtain energy.
- In conditions when lactose is the only source of carbon in the environment, the bacteria synthesises the β-galactosidase enzyme that breaks down the lactose into glucose and galactose.
- β-galactosidase (along with permease and transacetylase) that is required for lactose utilization is part of the lac operon.
- The lac operon consists of following gene:
- ‘i’ gene (inhibitor) : It codes for repressor of lac operon.
- ‘z’ gene (structural gene): It codes for β-galactosidase.
- ‘y’ gene (structural gene): It codes for permease, which increases the permeability of cell to lactose.
- ‘a’ gene (structural gene): It codes for transacetylase.
- Inducer: lactose acts as inducer. It regulates switching on and off of the operon.
Lac operon: in Absence of Inducer
- The repressor is synthesised all-the-time from the ‘i’ gene.
- The repressor protein binds to the operator region and prevents RNA polymerase from transcribing the operon.
- In this condition the lactose cannot be metabolised (as gene z, y and a are not expressed).
Lac operon: in presence of Inducer
- The inducer, allolactose (an alternate form of lactose), when present binds to the repressor and inactivated it.
- This inactivated repressor is unable to inactivate RNA polymerase enzyme and z, y, and a genes synthesise their respective mRNA, which in turn gets translated to form β-galactosidase, permease, and transacetylase.
- In presence of all these enzymes, the metabolism of lactose proceeds in a normal manner.
- This kind of regulation of lac operon is referred to as negative regulation.
Human Genome Project
- Joint venture of US department of energy and National Institute of Health (NIH); later joined by Welcome Trust (UK)
- Launched in 1990, completed in 2003
- This project worked towards the determination of complete DNA sequence of humans.
- Human genome (genome refers to the total genes that are present in a human being) contains 3 × 109 base pairs.
- If the obtained sequences were to be stored in typed form in books, and if each page of the book contained 1000 letters and each book contained 1000 pages, then 3300 such books would be required to store the information of DNA sequence from a single human cell.
- This creates problem for the storage and retrieval, and analysis of data.
- To solve this problem high speed computational devices were used. (A branch of biology called, bioinformatics developed).
Goals of HGP
- Some of the important goals of HGP were as follows:
- Identify all the approximately 20,000-25,000 genes in human DNA;
- Determine the sequences of the 3 billion chemical base pairs that make up human DNA;
- Store this information in databases;
- Improve tools for data analysis;
- Transfer related technologies to other sectors, such as industries;
- Address the ethical, legal, and social issues (ELSI) that may arise from the project.
- Genomes of many non-human models such as bacteria, yeast, Caenorhabditiselegans, Drosophila, plants (rice and Arabidopsis) have also been sequenced.
Methodologies used
- Two methods: identifying ESTs (Expressed sequence Tags) and sequence annotation
- ESTs: As the name suggests, this refers to the part of DNA that is expressed, i.e. transcribed, as mRNA and translated into proteins thereafter. It basically focuses on sequencing the part denoting a gene.
- Annotation: In this approach, entire genome (coding + non-coding) is sequenced and later on function is assigned to each region in the genome.
Genome Sequencing
- DNA from the cells is isolated and is randomly broken into fragments of smaller sizes.
- These fragments are cloned into suitable host using vectors.
- Cloned fragments amplify in the host. Amplification facilitates an easy sequencing.
- Common vectors used: BAC (Bacterial artificial chromosomes) and YAC (Yeast artificial chromosomes)
- Common hosts: Bacteria and yeasts
- Automated sequencers are used to sequence these smaller fragments (Sanger sequencing).
- The sequences so obtained are arranged based on overlapping regions within them (alignment).
- Alignment of the sequences is also done automatically by computer programs.
- Then these sequences are annotated and assigned to each chromosome.
Genetic and physical maps on Genome
- This was generated using information on polymorphism of restriction endonuclease recognition sites, and some repetitive DNA sequences known as microsatellites.
Salient Features of Human Genome
- The human genome contains 3164.7 million nucleotide bases.
- The average gene consists of 3000 bases.
- The total number of genes is estimated at 30,000.
- The functions are unknown for over 50 % of the discovered genes.
- Less than 2% of the genome codes for proteins.
- Repeated sequences make up very large portion of the human genome.
- Repetitive sequences that are repeated many times thought to have no direct coding functions, but they shed light on chromosome structure, dynamics and evolution.
- Chromosome 1 has most genes (2968), and the Y has the fewest (231).
- Scientists have identified about 1.4 million locations where single-base DNA differences (SNPs – single nucleotide polymorphism) occur in humans.
DNA Fingerprinting
- DNA fingerprinting is a method employed to compare the DNA sequences of any two individuals.
- Of the total base sequence present in humans, 99.9% in all human beings are identical. The remaining 0.1% differs from person to person and makes every individual unique.
- It is a really difficult and time-consuming task to sequence and compare all 3 × 109 bases in two individuals. So, instead of considering the entire genome, certain specific regions called repetitive DNA sequences are used for comparative study.
Basics of DNA fingerprinting
- In a density gradient centrifugation of bulk genomic DNA, most of the DNA formed a major peak, but the rest of the DNA formed a smaller peak called the Satellite DNA.
- Satellite DNA can be categorised as mini- or micro- depending on the following:
- Base composition (A:T and G:C rich)
- Length of segment
- Number of repetitive units
- These sequences do not code for any protein.
- Polymorphism is variation at genetic level that arises due to mutation.
- Satellite DNA show high degree of polymorphism and form the basis of DNA fingerprinting.
- As the polymorphisms are inheritable from parents to children, DNA fingerprinting is the basis of paternity testing, in case of disputes.
- Polymorphisms arise normally in the non-coding sequences because a mutation in non-coding sequences does not affect an individual’s reproductive ability.
Technique of DNA fingerprinting
- The technique was developed by Alec Jeffreys.
- Variable Number of Tandem Repeats (VNTR) is a type of satellite DNA that shows high degree of polymorphism.
- The various steps involved are as follows:
- Isolation of the DNA
- Digestion of the DNA with the help of restriction endonuclease (RE).
- The digestion with RE creates DNA fragments which are then separated by gel electrophoresis.
- The separated DNA fragments are blotted (immobilised) onto synthetic membranes, such as nitrocellulose or nylon.
- Immobilised fragments are hybridised with a VNTR probe.
- Detection of hybridised DNA fragments by autoradiography.
- VNTRs vary in size from 0.1 to 20 kb.
- The autoradiogram, after hybridization with VNTR probe gives many bands of different sizes.
- These bands give a characteristic pattern for an individual DNA.
- They are different in each individual, except identical twins.
Application:
- Test of paternity.
- Identification of the criminals.
- Population diversity determination.
- Determination of genetic diversity.
*********************

these notes are really helpful ……
Yes
Super notes sir #even following these notes for neet
This is what the perfect website that all students want to learn..
Thank you for your constructive comment.
I need the pdf of this ….not the online version..plz help
Dear Manisha, the PDF file is available for download. Please check at the end of the page for printer friendly PDF version.
Sir the pdf is unavailable
https://cbsebiology4u.blogspot.com/2021/03/biology-study-notes-cbse-class-12.html
You will get the files here at the end of the page.
It’s really a help full notes….
Thanks.
One of the best notes I have seen.
just wow!!!!!!
One of the best notes i have ever seen
Qbest notes..
very helpfull..
Nice. It ‘s Very helpful to me
very well written and upto the point … nothing less nothing more.. thank you so much for providing us with such wonderful notes….great job!!!…
Thank you…
Oh god thank you so much.i am preparing for qualifying neet In a month only thing I’m studying is biology from scratch as I am a US student to join med in India and this notes is a wow…it’s so helpful…🙏
Glad. You liked it. All the Best for the NEET.
best notes for most challenging chapter ,thank you sir for your divine work
Thank you Divya.
Thanku somuch it help me alot in making my notes and it is easy to understand …….thanku
Thanks
Thank you
best notes for all and easily understandable
Thank you
thanks
LITERALLY the best notes by far. These are so to the point notes, i have my board exam amd i prepared with this, its so sufficient & satisfactorily best!
Don’t have enough words to appreciate you Ramakant Sir. Thanks Alot 😊 Keep going sir.
Thank you dear.
Best notes of this chapter… approved by : Kim Namjoon
Sir it was amazing and really helpful.
This is only the website that covers all the topics with the great points you have written.
Please bio technology k notes bhi bbna legiye plzzzz.
Thnx…..
Chapter 11 Uploaded.