Biotechnology: Principles and Processes Class 12 Notes (Download pdf)
Table of Contents
Processes of Recombinant DNA technology
- Biotechnology – Field of biology that deals with the use of live organisms or biological systems to make products or processes that are useful to human beings.
- Biotechnology as per European Federation of Biotechnology (EFB) – The integration of natural science and organisms, cells, parts thereof, and molecular analogues for products and services.
❯ Principles of Biotechnology
✼ Genetic Engineering:
- Technique to modify genetic material.
- It allows the introduction of these modified genetic material into the host organisms.
- Can change the phenotype of the organism.
✼ Bioprocess Engineering:
- Maintenance of sterile condition to enable the growth of only desired microbes.
- Sterile: Free from microbial contamination.
- Prevents the growth of undesirable microbes.
❯ Some Basic Concepts
- What happens when a DNA molecule is somehow introduced into a host cell?
- The DNA cannot replicate within the host cell.
- Most probably it will be degraded.
- How can you make the foreign DNA multiply or make copies of itself within the host cell?
- The foreign DNA must become part of the host organism’s genome by integrating with it.
- Genome: All the genetic material of an organism.
- The foreign DNA must become part of the host organism’s genome by integrating with it.
- The foreign DNA now gets multiplied along with the host chromosome as the host chromosome contains the origin of replication.
- The origin of replication is essential to initiate the process of DNA replication.
- This process is called the cloning or making multiple identical copies of any template DNA.
✼ Construction and Cloning of the first artificial rDNA molecule:
- Scientists: Stanley Cohen and Herbert Boyer (1972)
- Native Plasmid of Salmonella typhimurium was linked to antibiotic resistant gene.
- Plasmid: Double stranded, autonomously replicating, extra chromosomal circular DNA that provides additional features to the bacterial cell if present.
- Antibiotic resistant gene: Gene that enables the bacteria to survive in a medium that contains antibiotics.
- Steps involved:
- Identification and cutting of a piece of DNA from a plasmid that provides resistance to antibiotics.
- The DNA was cut at a specific position.
- Enzyme involved: Restriction Enzymes – Molecular scissors.
- The cut piece of DNA was linked to the native plasmid of S. typhimurium.
- Enzyme involved: DNA Ligase
- This modified circular DNA is a recombinant DNA (rDNA) and acts as a vector.
- Vector – carrier of foreign DNA/gene of interest.
- rDNA – A DNA molecule that has been modified and may have a foreign DNA segment.
- Upon introduction of this rDNA into Escherichia coli bacteria, the rDNA was able to replicate within the new host (E. coli) and multiply in number using the new host’s DNA polymerase enzyme.
- As the original antibiotic resistant gene has been multiplied within the E. coli host, this process is referred to as the cloning of the antibiotic resistance gene in E. coli.
- Identification and cutting of a piece of DNA from a plasmid that provides resistance to antibiotics.
Three basic steps in genetically modifying an organism
- Identification of DNA with desirable genes;
- Introduction of the identified DNA into the host;
- Maintenance of introduced DNA in the host and transfer of the DNA to its progeny.
TOOLS OF RECOMBINANT DNA TECHNOLOGY
❯ Restriction Enzymes
- 1963 : Two enzymes were discovered that could restrict the growth of bacteriophage in E. coli.
- One enzyme added methyl group to DNA.
- The other enzyme could cut the DNA- restriction endonuclease.
- The first identified restriction endonuclease is Hind II.
- Restriction enzymes comes under Nucleases (larger class of enzyme).
- Two types of nuclease
- Exonuclease
- Removes nucleotides from the periphery/ends of the DNA.
- Endonuclease
- It cuts the DNA at specific positions away from the periphery within the DNA.
- Exonuclease
- Two types of nuclease
- The restriction endonuclease cuts the DNA at a specific position by recognizing a specific sequence of DNA referred to as recognition sequence.
- Each restriction endonuclease recognizes a specific palindromic nucleotide sequences in the DNA.
- Palindrome in DNA is a sequence of base pairs that reads same on the two strands when orientation of reading is kept the same.
- For example, the following sequences reads the same on the two strands in 5′ → 3′ direction. This is also true if read in the 3′ → 5′ direction.

- Each restriction endonuclease recognizes a specific palindromic nucleotide sequences in the DNA.
- Upon identification of the recognition sequence the RE binds with the DNA and cuts each of the two strands of the DNA double helix at specific points in their sugar-phosphate backbones.
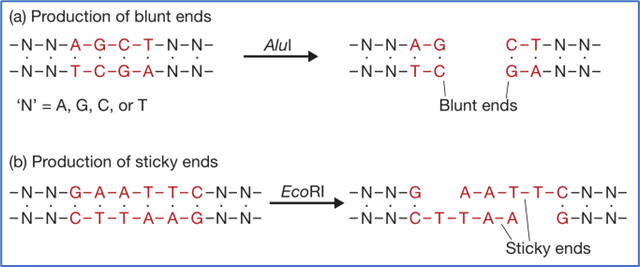
Blunt end and Sticky end
- Many RE make a simple double-stranded cut in the middle of the recognition sequence resulting in a blunt end.
- Examples of RE producing blunt end: PvuII and AluI.
- In other RE the two DNA strands are not cut at exactly the same position. Instead the cut is staggered, usually by two or four nucleotides, so that the resulting DNA fragments have short single-stranded overhangs at each end.
- These are called sticky ends, as base pairing between them can stick the DNA molecule back together again.
- This stickiness of the ends facilitates the action of the enzyme DNA ligase.
Nomenclature of Restriction Enzymes
- Rules of nomenclature of RE :
- The first letter of the name comes from the genus name.
- The second two letters come from the species of the prokaryotic cell from which they were isolated
- Following the three letters, the strain is sometimes represented.
- Following the strain, the Roman numbers following the names indicate the order in which the enzymes were isolated from that strain of bacteria.
- Examples: (need to remember the EcoRI nomenclature only)
| Enzymes Name | Genus name of Source organism | Species name of Source organism | Strain | Order of isolation from the bacteria strain |
| EcoRI | Escherichia | coli | RY 13 | I |
| HindII | Haemophilus | influenzae | d | II |
| HindIII | Haemophilus | influenzae | d | III |
| BamHI | Bacillus | amyloliquefaciens | H | I |
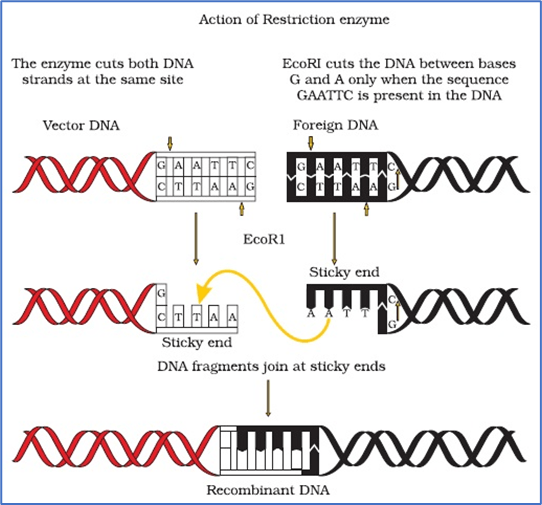
Steps in formation of recombinant DNA by action of restriction endonuclease enzyme – EcoRI
Recombinant DNA (rDNA)
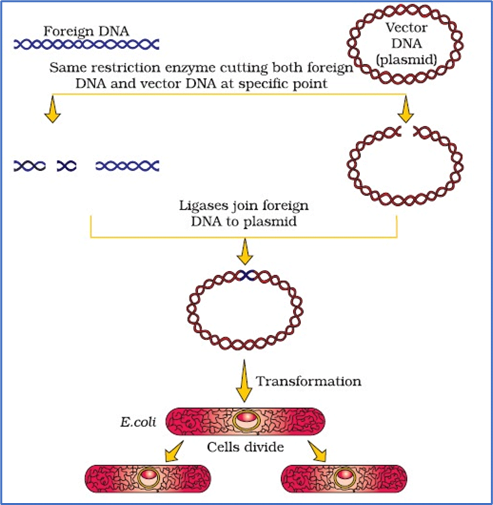
- RE are used in genetic engineering to form ‘recombinant’ molecules of DNA, which are composed of DNA from different sources/genomes.
- When cut by the same restriction enzyme, the resultant DNA fragments have the same kind of ‘sticky-ends’ and, these can be joined together (end-to-end) using DNA ligases.
- Unless one cuts the vector and the source DNA with the same restriction enzyme, the recombinant vector molecule cannot be created.
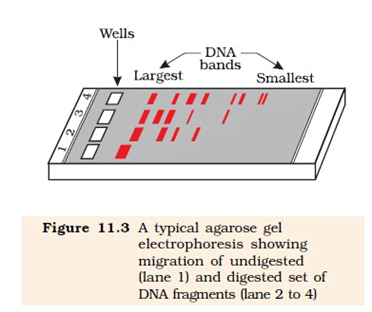
✼ Agarose Gel Electrophoresis
- Used for the separation and isolation of DNA fragments that is formed after a restriction digestion.
- Gel electrophoresis separates DNA molecules according to their size.
- DNA molecules are negatively charged.
- due to the presence of phosphate moiety.
- DNA molecules can be separated by forcing them to move towards anode under the influence of electric field through a medium/matrix.
- anode- positive pole.
- Matrix used: Agarose
- Agarose is extracted from sea weeds.
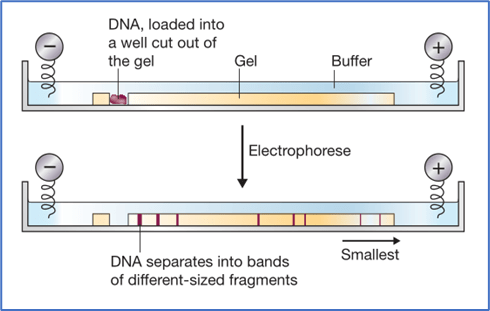
- The DNA fragments separate (resolve) according to their size through sieving effect provided by the agarose gel.
- Smaller DNA fragments: Moves the farthest.
- Larger DNA fragments: Moves the least and typically will be nearer to the loading wells of the agarose gel.
✼Visualization of separated DNA fragments
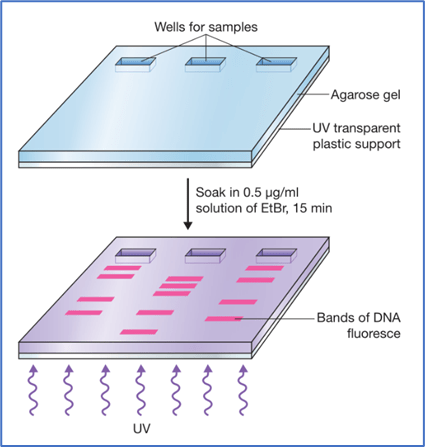
- DNA is not visible under normal light without staining with a compound that makes the DNA visible.
- Ethidium bromide (EtBr) is used for staining the DNA in the agarose gel.
- The different DNA bands in the gel is clearly visible under ultraviolet light after staining with EtBr.
- This procedure is very hazardous because ethidium bromide is a powerful mutagen.
- The DNA bands appear as bright orange colored bands.
✼Elution
- The process of cutting out the required separated bands of DNA from the agarose gel and extraction of the DNA from the gel piece after the gel electrophoresis is called elution.
- This purified DNA fragments then is used in constructing recombinant DNA by joining them with cloning vectors.
To continue reading select Click Here.
